Ligand c, with a percent occupancy of 30% at 1 μm Ligand d, with a percent occupancy of 80% at 10 nm c.
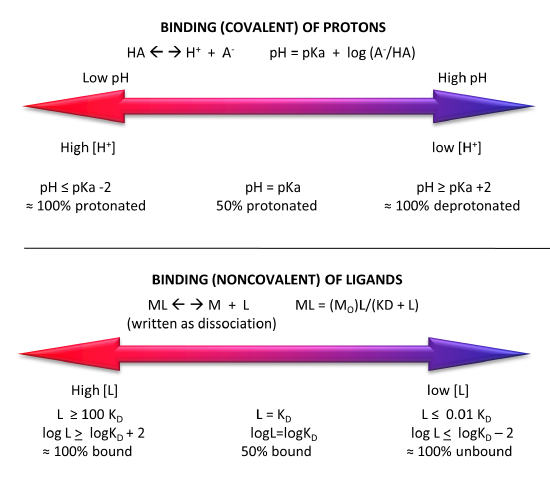
Which Ligand Binds Tightest. (c) ligand c, with a percent occupancy of 30% at one micromolar. Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other. When the immobilized ligand binds an analyte in solution, this local refractive index increases in direct proportion to the number of molecules bound to the sensor, making the refractive index shift equivalent to a change in mass. Ligand b, with a dissociation constant (𝐾dkd) of 10−3 m10−3 m.
 A) Chemical Structures Of Selected Pahs: Tightest Binding Guest… | Download Scientific Diagram From researchgate.net
A) Chemical Structures Of Selected Pahs: Tightest Binding Guest… | Download Scientific Diagram From researchgate.net
Related Post A) Chemical Structures Of Selected Pahs: Tightest Binding Guest… | Download Scientific Diagram :
If you add the central ion to a solution of the tighter binding ligand first, some of the ions will be occupied by multiple strong ligands, since the relative concentrations of the two will favor the ligand at first. Nm is (10�m), um is (10°m) and mm is (103m) a. In other words, the value represents the amount of inhibitor needed to. Ligand a, with a dissociation constant (𝐾dkd) of 10−9 m10−9 m.
C) ligand c, with a percent occupancy of 30% at 1 um.
(c) ligand c, with a percent occupancy of 30% at one micromolar. Ligand c, with a percent occupancy of 30% at 1 μm d. In other words, the value represents the amount of inhibitor needed to. Pwr has lower sensitivity than spr with regard to refractive index, thickness and mass parameters. It often, but not always, involves some chemical bonding. One bacterial protein called streptavidin binds to the biotin ligand extremely tightly.
 Source: chegg.com
Source: chegg.com
Ligand a, with a dissociation constant (𝐾dkd) of 10−9 m10−9 m. The ic50 value represents the concentration at which 50% of maximum possible inhibition occurs for a process. It is formed when atoms or molecules bind together by sharing of electrons.
 Source: slideserve.com
Source: slideserve.com
Ligand d, with a percent occupancy of 80% at 10 nm It often, but not always, involves some chemical bonding. (d) ligand d, with a per
 Source: nature.com
Source: nature.com
However, if the initial binding of the ligand results in a. The method described herein not only picks out the tightest binding ligands, it can also be used to detect ligands with. One bacterial protein called streptavidin binds to the biotin ligand extremely tightly.
 Source: naijaeduinfo.com
Source: naijaeduinfo.com
In addition, the limited dynamic range of visual readouts of gels that are often used to evaluate. Ligand d, with a percent occupancy of 80% at 10 nm c. Ligand c, with a percent occupancy of 30% at 1 μm
 Source: chegg.com
Source: chegg.com
Ligand d, with a percent occupancy of 80% at 10 nm. This effect is of positive cooperativity. Ligand d, with a percent occupancy of 80% at 10 nm.
 Source: quizlet.com
Source: quizlet.com
To answer this item, we must take note that the ligand that binds the tightest is the one with the lowest dissociation constant, kd. Therefore we can conclude that protein a binds ligand tighter at the midpoint of the binding curve. Learn this topic by watching enzymes and.
 Source: researchgate.net
Source: researchgate.net
This effect is of positive cooperativity. Ligand b, with a dissociation constant ( 𝐾d ) of 10−3 m d. If you add the central ion to a solution of the tighter binding ligand first, some of the ions will be occupied by multiple strong ligands, since the relative concentrations of the two will favor the ligand at first.
 Source: quizlet.com
Source: quizlet.com
Ligand b, with a dissociation constant ( 𝐾d ) of 10−3 m d. Kd�s for both a and b are already given so, we only need to solve kds for c and d. Ligand c, with a percent occupancy of 30% at 1 microm d.
 Source: chegg.com
Source: chegg.com
Learn this topic by watching enzymes and. Ligand binding changes amplitude, position and width of reflected lights. (c) ligand c, with a percent occupancy of 30% at one micromolar.
 Source: bio.libretexts.org
Source: bio.libretexts.org
Ligand c, with a percent occupancy of 30% at 1 μm Therefore we can conclude that protein a binds ligand tighter at the midpoint of the binding curve. The ic50 value represents the concentration at which 50% of maximum possible inhibition occurs for a process.
 Source: researchgate.net
Source: researchgate.net
It helps to know which ligand binds the tightest. C) ligand c, with a percent occupancy of 30% at 1 um. Kd= [p] [l]/ [pl] (p=protein, l=ligand) fraction of bonding = [l] / [l] + kd.
 Source: chemistry-europe.onlinelibrary.wiley.com
Source: chemistry-europe.onlinelibrary.wiley.com
Ligand b, with a dissociation constant ( 𝐾d ) of 10−3 m d. In some cases, the associations can be quite strong—for example, the. It often, but not always, involves some chemical bonding.
The ic50 value represents the concentration at which 50% of maximum possible inhibition occurs for a process. Ligand a, with a dissociation constant ( kd ) of 10−9 m ligand d, with a percent occupancy of 80% at 10 nm ligand b, with a dissociation constant ( kd ) of 10−3 m ligand c, with a percent occupancy of 30% at 1 μm In addition, the limited dynamic range of visual readouts of gels that are often used to evaluate.
 Source: study.com
Source: study.com
(c) ligand c, with a percent occupancy of 30% at one micromolar. (d) ligand d, with a per One bacterial protein called streptavidin binds to the biotin ligand extremely tightly.
 Source: numerade.com
Source: numerade.com
We�d like to design proteins that can bind ligands just as tightly, but this is a difficult protein design challenge. Pwr has lower sensitivity than spr with regard to refractive index, thickness and mass parameters. However, if the initial binding of the ligand results in a.

The method described herein not only picks out the tightest binding ligands, it can also be used to detect ligands with. Ligand a, with a dissociation constant ( kd ) of 10−9 m ligand d, with a percent occupancy of 80% at 10 nm ligand b, with a dissociation constant ( kd ) of 10−3 m ligand c, with a percent occupancy of 30% at 1 μm Ligand c, with a percent occupancy of 30% at 1 μm
 Source: quizlet.com
Source: quizlet.com
Pwr has lower sensitivity than spr with regard to refractive index, thickness and mass parameters. Ligand d, with a percent occupancy of 80% at 10 nm We�d like to design proteins that can bind ligands just as tightly, but this is a difficult protein design challenge.
 Source: numerade.com
Source: numerade.com
Ligand a, with a dissociation constant ( kd ) of 10−9 m ligand d, with a percent occupancy of 80% at 10 nm ligand b, with a dissociation constant ( kd ) of 10−3 m ligand c, with a percent occupancy of 30% at 1 μm Kd= [p] [l]/ [pl] (p=protein, l=ligand) fraction of bonding = [l] / [l] + kd. Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other.
 Source: chegg.com
Source: chegg.com
Ligand c, with a percent occupancy of 30% at 1 μm When the immobilized ligand binds an analyte in solution, this local refractive index increases in direct proportion to the number of molecules bound to the sensor, making the refractive index shift equivalent to a change in mass. Learn this topic by watching enzymes and.
 Source: chegg.com
Source: chegg.com
Ligand b, with a dissociation constant (𝐾dkd) of 10−3 m10−3 m. Pwr has lower sensitivity than spr with regard to refractive index, thickness and mass parameters. Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other.
Also Read :